zeligei-eihier¶
Wakefield’s Hierarchical Ecological Inference Model
Syntax¶
The EI models accept several different formula syntaxes. If 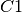 and are the column totals, and
and are the column totals, and 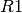 and are the row totals, and is the total in unit
and are the row totals, and is the total in unit 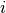 , then the formula can be expressed with just one row and one column, with the totals provided separately as:
, then the formula can be expressed with just one row and one column, with the totals provided separately as:
z.out <- zelig(C1 ~ R1, N=N, data=data)
The argument N can be either a numeric vector of the total in each i-th unit, or the character name of a variable in the dataset that contains these values.
Or with both rows and columns coupled together, and omitting the totals:
z.out <- zelig( cbind(C1,C2) ~ cbind(R1,R2), data=data)
Additionally, if C1, C2, R1, R2 are percentages rather than counts, then either formula method above is acceptable, however, N must always be provided.
First load packages:
library("Zelig")
library("ZeligEI")
Here is an example of all the syntax for the analysis using the first syntax method, and the direct use of the reference classes:
z5 <- zeihier$new()
z5$zelig(C1 ~ R1, N=myN, weights = w, data = myData)
z5$setx()
z5$sim()
With the Zelig 4 compatibility wrappers this looks like:
z.out <- zelig(C1 ~ R1, N=N, model = "eihier", weights = w, data = myData)
x.out <- setx(z.out)
s.out <- sim(z.out, x = x.out)
Additional Inputs¶
In addition, Zelig accepts the following additional inputs for eihier to monitor the convergence of the Markov chain:
- burnin: number of the initial MCMC iterations to be discarded (defaults to 5,000).
- mcmc: number of the MCMC iterations after burnin (defaults to 50,000).
- thin: thinning interval for the Markov chain. Only every thin-th draw from the Markov chain is kept. The value of mcmc must be divisible by this value. The default value is 1.
- verbose: defaults to FALSE. If TRUE, the progress of the sampler (every 10%) is printed to the screen.
- seed: seed for the random number generator. The default is NA which corresponds to a random seed of 12345.
The model also accepts the following additional arguments to specify prior parameters used in the model:
- m0: prior mean of (defaults to 0).
- M0: prior variance of (defaults to 2.287656).
- m1: prior mean of (defaults to 0).
- M1: prior variance of (defaults to 2.287656).
- a0: is the shape parameter for the Inverse Gamma prior on . The default is 0.825.
- b0: is the scale parameter for the Inverse Gamma prior on . The default is 0.0105.
- a1: is the shape parameter for the Inverse Gamma prior on . The default is 0.825.
- b1: is the scale parameter for the Inverse Gamma prior on . The default is 0.0105.
Users may wish to refer to help(MCMCdynamicEI) for more options.
Examples¶
We’ll use a dataset from the ei package, of black and non-black turnout in 141 precincts.
library("ei", quietly=TRUE)
##
## Attaching package: 'coda'
## The following object is masked from 'package:arm':
##
## traceplot
data(sample)
Here is the model estimated in Zelig.
z.out <- zeihier$new()
z.out$zelig(t~x, N="n", data=sample)
summary(z.out)
## Model:
##
## Iterations = 5001:55000
## Thinning interval = 1
## Number of chains = 1
## Sample size per chain = 50000
##
## 1. Empirical mean and standard deviation for each variable,
## plus standard error of the mean:
##
## Mean SD Naive SE Time-series SE
## p0table1 0.071436 0.078023 3.49e-04 6.48e-03
## p0table2 0.080973 0.114551 5.12e-04 1.30e-02
## p0table3 0.072274 0.082796 3.70e-04 6.58e-03
## p0table4 0.077294 0.103491 4.63e-04 8.86e-03
## p0table5 0.077767 0.104512 4.67e-04 9.09e-03
## p0table6 0.074752 0.093577 4.18e-04 7.38e-03
## p0table7 0.078859 0.108135 4.84e-04 1.17e-02
## p0table8 0.079053 0.109479 4.90e-04 9.87e-03
## p0table9 0.071804 0.079629 3.56e-04 6.82e-03
## p0table10 0.071656 0.079061 3.54e-04 6.30e-03
## p0table11 0.071977 0.081226 3.63e-04 5.80e-03
## p0table12 0.069398 0.069386 3.10e-04 5.63e-03
## p0table13 0.073864 0.088924 3.98e-04 7.63e-03
## p0table14 0.075628 0.096575 4.32e-04 7.85e-03
## p0table15 0.069987 0.071474 3.20e-04 5.84e-03
## p0table16 0.074841 0.093829 4.20e-04 6.94e-03
## p0table17 0.081995 0.117027 5.23e-04 1.28e-02
## p0table18 0.080602 0.114533 5.12e-04 1.17e-02
## p0table19 0.080430 0.111818 5.00e-04 1.15e-02
## p0table20 0.079105 0.107834 4.82e-04 9.98e-03
## p0table21 0.078316 0.106679 4.77e-04 1.02e-02
## p0table22 0.069781 0.069642 3.11e-04 5.93e-03
## p0table23 0.077192 0.101955 4.56e-04 9.52e-03
## p0table24 0.070463 0.072994 3.26e-04 6.04e-03
## p0table25 0.081716 0.116677 5.22e-04 1.25e-02
## p0table26 0.076926 0.100804 4.51e-04 8.86e-03
## p0table27 0.073473 0.087508 3.91e-04 7.48e-03
## p0table28 0.075491 0.096164 4.30e-04 7.90e-03
## p0table29 0.080572 0.113409 5.07e-04 1.28e-02
## p0table30 0.071206 0.076504 3.42e-04 5.93e-03
## p0table31 0.074835 0.092900 4.15e-04 7.49e-03
## p0table32 0.078493 0.107157 4.79e-04 1.14e-02
## p0table33 0.080990 0.114045 5.10e-04 1.19e-02
## p0table34 0.069615 0.069599 3.11e-04 5.82e-03
## p0table35 0.075882 0.097188 4.35e-04 8.55e-03
## p0table36 0.077248 0.103221 4.62e-04 9.18e-03
## p0table37 0.070617 0.074170 3.32e-04 5.81e-03
## p0table38 0.082013 0.118051 5.28e-04 1.19e-02
## p0table39 0.081788 0.115987 5.19e-04 1.33e-02
## p0table40 0.078940 0.108788 4.87e-04 1.11e-02
## p0table41 0.074680 0.093349 4.17e-04 8.07e-03
## p0table42 0.075691 0.096700 4.32e-04 8.36e-03
## p0table43 0.072316 0.082915 3.71e-04 7.18e-03
## p0table44 0.080306 0.112131 5.01e-04 1.16e-02
## p0table45 0.078590 0.106996 4.78e-04 1.08e-02
## p0table46 0.077231 0.102901 4.60e-04 8.49e-03
## p0table47 0.081068 0.114822 5.14e-04 1.12e-02
## p0table48 0.074158 0.089863 4.02e-04 7.28e-03
## p0table49 0.080546 0.112917 5.05e-04 1.17e-02
## p0table50 0.078429 0.107117 4.79e-04 9.64e-03
## p0table51 0.073102 0.086626 3.87e-04 5.95e-03
## p0table52 0.079114 0.109408 4.89e-04 1.01e-02
## p0table53 0.079457 0.110799 4.96e-04 1.04e-02
## p0table54 0.076338 0.099930 4.47e-04 8.62e-03
## p0table55 0.070931 0.075591 3.38e-04 6.17e-03
## p0table56 0.075460 0.096036 4.29e-04 8.03e-03
## p0table57 0.070824 0.075291 3.37e-04 5.92e-03
## p0table58 0.080882 0.114202 5.11e-04 1.23e-02
## p0table59 0.078994 0.109557 4.90e-04 1.08e-02
## p0table60 0.070305 0.072105 3.22e-04 5.81e-03
## p0table61 0.081349 0.113797 5.09e-04 1.19e-02
## p0table62 0.080429 0.112856 5.05e-04 1.14e-02
## p0table63 0.081051 0.114635 5.13e-04 1.22e-02
## p0table64 0.073916 0.089371 4.00e-04 7.04e-03
## p0table65 0.078977 0.108073 4.83e-04 1.13e-02
## p0table66 0.078398 0.106532 4.76e-04 1.07e-02
## p0table67 0.075188 0.095212 4.26e-04 7.72e-03
## p0table68 0.078378 0.106186 4.75e-04 9.46e-03
## p0table69 0.082377 0.116754 5.22e-04 1.26e-02
## p0table70 0.079149 0.107455 4.81e-04 9.74e-03
## p0table71 0.082340 0.118188 5.29e-04 1.18e-02
## p0table72 0.081234 0.114100 5.10e-04 1.27e-02
## p0table73 0.071516 0.078784 3.52e-04 6.94e-03
## p0table74 0.073846 0.089262 3.99e-04 7.35e-03
## p0table75 0.071013 0.075761 3.39e-04 5.63e-03
## p1table1 0.000454 0.000107 4.80e-07 4.85e-06
## p1table2 0.000466 0.000111 4.98e-07 4.92e-06
## p1table3 0.000454 0.000107 4.77e-07 5.09e-06
## p1table4 0.000458 0.000107 4.79e-07 5.19e-06
## p1table5 0.000458 0.000108 4.83e-07 5.08e-06
## p1table6 0.000454 0.000109 4.87e-07 5.07e-06
## p1table7 0.000461 0.000108 4.84e-07 4.95e-06
## p1table8 0.000461 0.000109 4.87e-07 5.00e-06
## p1table9 0.000454 0.000107 4.80e-07 4.96e-06
## p1table10 0.000455 0.000108 4.82e-07 5.10e-06
## p1table11 0.000453 0.000107 4.80e-07 5.21e-06
## p1table12 0.000454 0.000108 4.83e-07 5.06e-06
## p1table13 0.000455 0.000108 4.83e-07 5.16e-06
## p1table14 0.000457 0.000109 4.86e-07 5.20e-06
## p1table15 0.000454 0.000107 4.80e-07 5.11e-06
## p1table16 0.000456 0.000108 4.84e-07 5.06e-06
## p1table17 0.000472 0.000113 5.06e-07 4.99e-06
## p1table18 0.000466 0.000112 5.01e-07 5.13e-06
## p1table19 0.000472 0.000115 5.13e-07 5.08e-06
## p1table20 0.000474 0.000114 5.08e-07 4.92e-06
## p1table21 0.000459 0.000108 4.84e-07 4.79e-06
## p1table22 0.000454 0.000106 4.75e-07 5.10e-06
## p1table23 0.000458 0.000108 4.82e-07 5.12e-06
## p1table24 0.000454 0.000107 4.80e-07 5.00e-06
## p1table25 0.000467 0.000112 5.02e-07 4.95e-06
## p1table26 0.000457 0.000107 4.81e-07 5.03e-06
## p1table27 0.000454 0.000107 4.80e-07 5.20e-06
## p1table28 0.000458 0.000109 4.88e-07 4.90e-06
## p1table29 0.000465 0.000112 5.02e-07 5.12e-06
## p1table30 0.000454 0.000106 4.75e-07 5.28e-06
## p1table31 0.000456 0.000108 4.82e-07 4.94e-06
## p1table32 0.000461 0.000110 4.93e-07 5.12e-06
## p1table33 0.000468 0.000113 5.05e-07 5.13e-06
## p1table34 0.000454 0.000109 4.86e-07 5.20e-06
## p1table35 0.000457 0.000108 4.83e-07 5.03e-06
## p1table36 0.000457 0.000108 4.84e-07 5.20e-06
## p1table37 0.000453 0.000108 4.82e-07 5.13e-06
## p1table38 0.000467 0.000111 4.97e-07 5.16e-06
## p1table39 0.000472 0.000113 5.05e-07 4.89e-06
## p1table40 0.000460 0.000108 4.83e-07 5.00e-06
## p1table41 0.000455 0.000107 4.78e-07 4.98e-06
## p1table42 0.000457 0.000108 4.84e-07 4.90e-06
## p1table43 0.000454 0.000107 4.80e-07 5.09e-06
## p1table44 0.000468 0.000113 5.05e-07 5.07e-06
## p1table45 0.000459 0.000109 4.86e-07 4.95e-06
## p1table46 0.000457 0.000108 4.83e-07 5.01e-06
## p1table47 0.000465 0.000110 4.92e-07 5.07e-06
## p1table48 0.000456 0.000107 4.77e-07 4.98e-06
## p1table49 0.000466 0.000111 4.96e-07 5.23e-06
## p1table50 0.000460 0.000108 4.85e-07 5.05e-06
## p1table51 0.000456 0.000109 4.86e-07 4.96e-06
## p1table52 0.000461 0.000109 4.88e-07 5.05e-06
## p1table53 0.000462 0.000110 4.90e-07 4.98e-06
## p1table54 0.000457 0.000109 4.88e-07 5.05e-06
## p1table55 0.000455 0.000107 4.80e-07 5.10e-06
## p1table56 0.000456 0.000107 4.79e-07 5.01e-06
## p1table57 0.000455 0.000108 4.84e-07 5.07e-06
## p1table58 0.000466 0.000111 4.98e-07 4.89e-06
## p1table59 0.000461 0.000109 4.89e-07 5.08e-06
## p1table60 0.000453 0.000107 4.77e-07 5.09e-06
## p1table61 0.000470 0.000110 4.93e-07 5.11e-06
## p1table62 0.000465 0.000110 4.94e-07 5.08e-06
## p1table63 0.000470 0.000111 4.98e-07 5.15e-06
## p1table64 0.000454 0.000107 4.79e-07 5.08e-06
## p1table65 0.000463 0.000109 4.88e-07 5.11e-06
## p1table66 0.000460 0.000108 4.83e-07 5.05e-06
## p1table67 0.000455 0.000107 4.77e-07 4.88e-06
## p1table68 0.000461 0.000108 4.83e-07 4.83e-06
## p1table69 0.000476 0.000117 5.24e-07 5.05e-06
## p1table70 0.000469 0.000110 4.94e-07 4.94e-06
## p1table71 0.000469 0.000112 5.01e-07 5.17e-06
## p1table72 0.000476 0.000113 5.03e-07 5.04e-06
## p1table73 0.000454 0.000108 4.81e-07 5.13e-06
## p1table74 0.000454 0.000107 4.81e-07 5.10e-06
## p1table75 0.000455 0.000108 4.81e-07 5.19e-06
## mu0 -2.868125 0.865837 3.87e-03 1.23e-01
## mu1 -7.710528 0.173314 7.75e-04 1.24e-02
## sigma^2.0 1.009668 8.796670 3.93e-02 5.43e-01
## sigma^2.1 0.028775 0.046034 2.06e-04 2.14e-03
##
## 2. Quantiles for each variable:
##
## 2.5% 25% 50% 75% 97.5%
## p0table1 0.007334 0.030204 0.051509 0.092133 0.226094
## p0table2 0.008097 0.030895 0.052737 0.095122 0.292241
## p0table3 0.007502 0.030150 0.051557 0.092805 0.231089
## p0table4 0.007910 0.030606 0.052071 0.093718 0.257752
## p0table5 0.007971 0.030606 0.052359 0.093965 0.261572
## p0table6 0.007620 0.030356 0.052062 0.093065 0.244019
## p0table7 0.007926 0.030713 0.052259 0.094266 0.273386
## p0table8 0.007970 0.030779 0.052440 0.094121 0.274642
## p0table9 0.007257 0.030334 0.051716 0.092626 0.227308
## p0table10 0.007537 0.030153 0.051768 0.092104 0.227463
## p0table11 0.007451 0.030096 0.051425 0.092026 0.230965
## p0table12 0.007169 0.029939 0.051315 0.091538 0.213278
## p0table13 0.007492 0.030427 0.051813 0.092691 0.239910
## p0table14 0.007636 0.030581 0.052016 0.093600 0.249649
## p0table15 0.007242 0.030020 0.051404 0.091806 0.217634
## p0table16 0.007657 0.030416 0.052003 0.092824 0.242830
## p0table17 0.008097 0.031081 0.053024 0.095444 0.309722
## p0table18 0.008051 0.030936 0.052688 0.094609 0.291766
## p0table19 0.008280 0.030961 0.052648 0.094895 0.293962
## p0table20 0.008094 0.030817 0.052635 0.094713 0.278270
## p0table21 0.008075 0.030885 0.052445 0.094243 0.267480
## p0table22 0.007296 0.030015 0.051508 0.091662 0.221939
## p0table23 0.007757 0.030734 0.052328 0.093928 0.261274
## p0table24 0.007258 0.029935 0.051521 0.092066 0.223657
## p0table25 0.008079 0.031107 0.052865 0.095553 0.303024
## p0table26 0.007873 0.030584 0.052147 0.094222 0.258724
## p0table27 0.007526 0.030259 0.051750 0.092659 0.239638
## p0table28 0.007758 0.030462 0.051984 0.093278 0.248120
## p0table29 0.008054 0.030978 0.052878 0.094555 0.289962
## p0table30 0.007254 0.030119 0.051597 0.092101 0.227370
## p0table31 0.007792 0.030304 0.051872 0.093012 0.244027
## p0table32 0.007980 0.030600 0.052328 0.094229 0.273628
## p0table33 0.008084 0.030985 0.052602 0.094811 0.300736
## p0table34 0.007172 0.029832 0.051317 0.091376 0.219639
## p0table35 0.007623 0.030497 0.052068 0.093480 0.253780
## p0table36 0.007839 0.030697 0.052147 0.093850 0.260327
## p0table37 0.007151 0.030019 0.051417 0.091948 0.224274
## p0table38 0.008273 0.031035 0.053008 0.095182 0.301218
## p0table39 0.008353 0.031098 0.053122 0.095737 0.303734
## p0table40 0.008026 0.030650 0.052495 0.094156 0.275872
## p0table41 0.007649 0.030240 0.051942 0.092952 0.242450
## p0table42 0.007680 0.030544 0.052058 0.093197 0.250878
## p0table43 0.007504 0.030245 0.051515 0.092581 0.230677
## p0table44 0.008228 0.031005 0.052816 0.095071 0.282990
## p0table45 0.007910 0.030595 0.052350 0.094379 0.276969
## p0table46 0.007894 0.030523 0.052147 0.094028 0.259708
## p0table47 0.008157 0.030940 0.052572 0.094787 0.302381
## p0table48 0.007643 0.030487 0.052053 0.093009 0.242739
## p0table49 0.008230 0.030957 0.052475 0.094702 0.292066
## p0table50 0.007826 0.030739 0.052351 0.093948 0.266893
## p0table51 0.007624 0.030256 0.051812 0.092627 0.233090
## p0table52 0.007982 0.030649 0.052569 0.094436 0.278852
## p0table53 0.008066 0.030950 0.052506 0.094342 0.277700
## p0table54 0.007876 0.030518 0.052162 0.093283 0.255370
## p0table55 0.007326 0.030157 0.051634 0.092050 0.227015
## p0table56 0.007762 0.030464 0.052131 0.093612 0.245558
## p0table57 0.007298 0.030116 0.051566 0.091641 0.224876
## p0table58 0.008182 0.030978 0.052802 0.095118 0.297408
## p0table59 0.008040 0.030558 0.052243 0.094072 0.273982
## p0table60 0.007315 0.030141 0.051519 0.092104 0.218726
## p0table61 0.008190 0.030829 0.052945 0.095143 0.304161
## p0table62 0.008176 0.030970 0.052811 0.095065 0.285795
## p0table63 0.008156 0.031006 0.052724 0.095181 0.294578
## p0table64 0.007454 0.030378 0.051875 0.092615 0.241226
## p0table65 0.008054 0.030624 0.052690 0.094049 0.277606
## p0table66 0.007879 0.030644 0.052349 0.094237 0.267574
## p0table67 0.007707 0.030365 0.051864 0.093303 0.244718
## p0table68 0.008098 0.030672 0.052327 0.094133 0.271527
## p0table69 0.008364 0.031162 0.053023 0.096031 0.317175
## p0table70 0.008105 0.030722 0.052648 0.094779 0.279647
## p0table71 0.008399 0.030891 0.052929 0.095464 0.315779
## p0table72 0.008149 0.030899 0.052934 0.095537 0.300193
## p0table73 0.007252 0.030127 0.051757 0.092043 0.226904
## p0table74 0.007707 0.030541 0.051831 0.092907 0.238765
## p0table75 0.007396 0.030001 0.051610 0.092243 0.226280
## p1table1 0.000258 0.000385 0.000449 0.000515 0.000681
## p1table2 0.000276 0.000394 0.000458 0.000526 0.000703
## p1table3 0.000260 0.000386 0.000449 0.000515 0.000678
## p1table4 0.000265 0.000389 0.000452 0.000518 0.000683
## p1table5 0.000264 0.000389 0.000452 0.000518 0.000688
## p1table6 0.000258 0.000384 0.000449 0.000516 0.000684
## p1table7 0.000270 0.000392 0.000454 0.000521 0.000686
## p1table8 0.000268 0.000391 0.000454 0.000521 0.000691
## p1table9 0.000258 0.000387 0.000450 0.000515 0.000680
## p1table10 0.000260 0.000386 0.000450 0.000516 0.000679
## p1table11 0.000259 0.000384 0.000448 0.000515 0.000677
## p1table12 0.000260 0.000386 0.000449 0.000515 0.000678
## p1table13 0.000259 0.000387 0.000450 0.000516 0.000681
## p1table14 0.000262 0.000388 0.000451 0.000517 0.000682
## p1table15 0.000260 0.000385 0.000449 0.000515 0.000680
## p1table16 0.000262 0.000387 0.000450 0.000516 0.000680
## p1table17 0.000283 0.000400 0.000463 0.000530 0.000717
## p1table18 0.000276 0.000395 0.000457 0.000524 0.000703
## p1table19 0.000282 0.000400 0.000463 0.000530 0.000721
## p1table20 0.000287 0.000402 0.000464 0.000532 0.000719
## p1table21 0.000266 0.000390 0.000453 0.000519 0.000691
## p1table22 0.000260 0.000385 0.000449 0.000515 0.000680
## p1table23 0.000263 0.000389 0.000452 0.000518 0.000686
## p1table24 0.000258 0.000386 0.000449 0.000515 0.000684
## p1table25 0.000276 0.000396 0.000459 0.000526 0.000703
## p1table26 0.000261 0.000389 0.000451 0.000518 0.000684
## p1table27 0.000259 0.000385 0.000449 0.000515 0.000680
## p1table28 0.000263 0.000389 0.000452 0.000518 0.000686
## p1table29 0.000273 0.000395 0.000456 0.000524 0.000703
## p1table30 0.000257 0.000385 0.000450 0.000516 0.000676
## p1table31 0.000262 0.000387 0.000450 0.000517 0.000683
## p1table32 0.000268 0.000391 0.000454 0.000521 0.000694
## p1table33 0.000277 0.000397 0.000460 0.000527 0.000713
## p1table34 0.000260 0.000386 0.000449 0.000516 0.000682
## p1table35 0.000264 0.000387 0.000451 0.000518 0.000684
## p1table36 0.000263 0.000388 0.000451 0.000518 0.000686
## p1table37 0.000258 0.000386 0.000448 0.000515 0.000679
## p1table38 0.000277 0.000396 0.000459 0.000526 0.000705
## p1table39 0.000283 0.000400 0.000463 0.000530 0.000714
## p1table40 0.000266 0.000390 0.000453 0.000520 0.000687
## p1table41 0.000260 0.000387 0.000450 0.000515 0.000681
## p1table42 0.000262 0.000388 0.000451 0.000517 0.000682
## p1table43 0.000259 0.000385 0.000448 0.000516 0.000679
## p1table44 0.000278 0.000397 0.000460 0.000527 0.000704
## p1table45 0.000265 0.000389 0.000452 0.000519 0.000687
## p1table46 0.000263 0.000388 0.000451 0.000518 0.000685
## p1table47 0.000274 0.000394 0.000458 0.000524 0.000699
## p1table48 0.000261 0.000387 0.000450 0.000516 0.000679
## p1table49 0.000277 0.000396 0.000458 0.000525 0.000704
## p1table50 0.000267 0.000391 0.000453 0.000520 0.000689
## p1table51 0.000259 0.000387 0.000450 0.000516 0.000682
## p1table52 0.000267 0.000391 0.000454 0.000521 0.000694
## p1table53 0.000269 0.000392 0.000455 0.000521 0.000693
## p1table54 0.000262 0.000387 0.000451 0.000518 0.000689
## p1table55 0.000260 0.000385 0.000450 0.000517 0.000680
## p1table56 0.000261 0.000387 0.000451 0.000517 0.000681
## p1table57 0.000256 0.000386 0.000449 0.000516 0.000681
## p1table58 0.000274 0.000395 0.000458 0.000525 0.000704
## p1table59 0.000268 0.000391 0.000455 0.000522 0.000694
## p1table60 0.000259 0.000385 0.000449 0.000515 0.000676
## p1table61 0.000283 0.000399 0.000461 0.000528 0.000710
## p1table62 0.000272 0.000395 0.000457 0.000524 0.000698
## p1table63 0.000280 0.000399 0.000461 0.000527 0.000710
## p1table64 0.000258 0.000386 0.000449 0.000516 0.000679
## p1table65 0.000274 0.000393 0.000456 0.000522 0.000696
## p1table66 0.000266 0.000391 0.000454 0.000520 0.000690
## p1table67 0.000264 0.000387 0.000450 0.000516 0.000680
## p1table68 0.000267 0.000391 0.000454 0.000521 0.000690
## p1table69 0.000288 0.000402 0.000465 0.000533 0.000725
## p1table70 0.000282 0.000398 0.000460 0.000528 0.000710
## p1table71 0.000280 0.000398 0.000460 0.000527 0.000712
## p1table72 0.000289 0.000403 0.000466 0.000534 0.000721
## p1table73 0.000260 0.000386 0.000449 0.000515 0.000676
## p1table74 0.000259 0.000386 0.000448 0.000516 0.000679
## p1table75 0.000260 0.000387 0.000449 0.000516 0.000678
## mu0 -4.675128 -3.410300 -2.885947 -2.265954 -1.219557
## mu1 -8.101791 -7.815823 -7.697711 -7.593974 -7.400586
## sigma^2.0 0.002072 0.007243 0.018905 0.064123 4.495299
## sigma^2.1 0.001870 0.005960 0.013131 0.031202 0.156413
##
## Next step: Use 'setx' method
You can check for convergence before summarizing the estimates with three diagnostic tests. See the section Diagnostics for Zelig Models for examples of the output with interpretation:
z.out$geweke.diag()
z.out$heidel.diag()
z.out$raftery.diag()
See also¶
The Wakefield hierarchical model is part of the MCMCpack package by Andrew Martin, Kevin Quinn, and Jong Hee Park. Advanced users may wish to refer to help(MCMChierEI) in the MCMCpack package.
Wakefield J (2004). “Ecological Inference for 2 x 2 Tables.” Journal of the Royal Statistical Society, Series A., 167 (3), pp. 385-445.
Martin AD, Quinn KM and Park JH (2011). “MCMCpack: Markov Chain Monte Carlo in R.” Journal of Statistical Software, 42 (9), pp. 22. <URL: http://www.jstatsoft.org/v42/i09/>.