Warning
ZeligMultilevel is currently under development. We are working on the model vignettes of the package.
zeligmultilevel-poissonmixed¶
poisson.mixed: Mixed effects poisson Regression
Use generalized multi-level linear regression if you have covariates that are grouped according to one or more classification factors. Poisson regression applies to dependent variables that represent the number of independent events that occur during a fixed period of time.
While generally called multi-level models in the social sciences, this class of models is often referred to as mixed-effects models in the statistics literature and as hierarchical models in a Bayesian setting. This general class of models consists of linear models that are expressed as a function of both fixed effects, parameters corresponding to an entire population or certain repeatable levels of experimental factors, and random effects, parameters corresponding to individual experimental units drawn at random from a population.
Syntax¶
z5 <- zpoissonmixed$new()
z5$zelig(formula= y ~ x1 + x2 + (z1 + z2 | g), weights = w, data = mydata)
z5$setx()
z5$sim()
With the Zelig 4 compatibility wrappers:
z.out <- zelig(formula= y ~ x1 + x2 + (z1 + z2 | g),
data = mydata, weights = w, model = "poisson.mixed")
x.out <- setx(z.out)
s.out <- sim(z.out, x = x.out)
Inputs¶
zelig() takes the following arguments for multi:
- formula: a two-sided linear formula object describing the systematic component of the model, with the response on the left of a operator and the fixed effects terms, separated by + operators, on the right. Any random effects terms are included with the notation (z1 + ... + zn | g) with z1 + ... + zn specifying the model for the random effects and g the grouping structure. Random intercept terms are included with the notation (1 | g).
Additional Inputs¶
Additionally, users may wish to refer to glmer in the package lme4 for more information, including control parameters for the estimation algorithm and their defaults.
Examples¶
Basic Example¶
Attach sample data:
data(homerun)
Estimate model:
z.out <- zelig(homeruns ~ player + (player - 1 | month),
data = homerun, model = "poisson.mixed")
## Warning in readLines(zeligmixedmodels): incomplete final line found on
## '/usr/lib64/R/library/ZeligMultilevel/JSON/zelig5mixedmodels.json'
## How to cite this model in Zelig:
## TBD. 2016.
## poisson.mixed:
## in Christine Choirat, James Honaker, Kosuke Imai, Gary King, and Olivia Lau,
## "Zelig: Everyone's Statistical Software," http://zeligproject.org/
Summarize regression coefficients and estimated variance of random effects:
summary(z.out)
## Model:
## Generalized linear mixed model fit by maximum likelihood (Laplace
## Approximation) [glmerMod]
## Family: poisson ( log )
## Formula: homeruns ~ player + (player - 1 | month)
## Data: homerun
##
## AIC BIC logLik deviance df.resid
## 544 562 -267 534 309
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -0.746 -0.667 -0.618 0.810 3.727
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## month playerMcGwire 0.0020 0.0447
## playerSosa 0.0674 0.2597 -1.00
## Number of obs: 314, groups: month, 7
##
## Fixed effects:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.795 0.121 -6.57 5e-11
## playerSosa -0.127 0.217 -0.59 0.56
##
## Correlation of Fixed Effects:
## (Intr)
## playerSosa -0.631
## Next step: Use 'setx' method
Set explanatory variables to their default values:
x.out <- setx(z.out)
## [1] "month"
Simulate draws using the default bootstrap method and view simulated quantities of interest:
##
## sim x :
## -----
## ev
## mean sd 50% 2.5% 97.5%
## [1,] 0.379 0.138 0.357 0.179 0.708
## pv
## mean sd 50% 2.5% 97.5%
## [1,] 0.378 0.634 0 0 2
s.out <- sim(z.out, x = x.out)
summary(s.out)
plot(s.out)
Graphs of Quantities of Interest for Poisson Mixed Model
Mixed effects Poisson Regression Model¶
Let be the number of independent events that occur during
a fixed time period, realized for observation in group
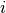 as , which takes any non-negative integer as its
value, for , .
as , which takes any non-negative integer as its
value, for , .
The stochastic component is described by a Poisson distribution with mean and variance parameter .
where
The -dimensional vector of random effects, , is restricted to be mean zero, and therefore is completely characterized by the variance covarance matrix , a symmetric positive semi-definite matrix.
The systematic component is
where is the array of known fixed effects explanatory variables,
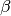 is the
-dimensional vector of fixed effects coefficients,
is the array of known
random effects explanatory variables and is the
-dimensional vector of random effects.
is the
-dimensional vector of fixed effects coefficients,
is the array of known
random effects explanatory variables and is the
-dimensional vector of random effects.
Quantities of Interest¶
The predicted values are draws from the poisson distribution defined by mean , for
given and and simulations of of
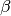 and from their posterior distributions. The
estimated variance covariance matrices are taken as correct and are
themselves not simulated.
and from their posterior distributions. The
estimated variance covariance matrices are taken as correct and are
themselves not simulated.The expected values is the mean of simulations of the stochastic component given draws of
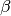 from its posterior:
from its posterior:The first difference is given by the difference in expected values, conditional on and , representing different values of the explanatory variables.
In conditional prediction models, the average predicted treatment effect for the treatment group is given by
where is a binary explanatory variable defining the treatment and control groups. Variation in the simulations is due to uncertainty in simulating , the counterfactual predicted value of for observations in the treatment group, under the assumption that everything stays the same except that the treatment indicator is switched to .
In conditional prediction models, the average expected treatment effect for the treatment group is given by
where is a binary explanatory variable defining the treatment and control groups. Variation in the simulations is due to uncertainty in simulating , the counterfactual expected value of for observations in the treatment group, under the assumption that everything stays the same except that the treatment indicator is switched to .
Output Values¶
The output of each Zelig command contains useful information which you may view. You may examine the available information in z.out by using getters (http://docs.zeligproject.org/en/latest/getters.html), see the fixed effect coefficients by using z.out$getcoef(), and a default summary of information through summary(z.out). Other elements available are listed below.
- From the zelig() output stored in summary(z.out), you may extract:
- fixef: numeric vector containing the conditional estimates of the fixed effects.
- ranef: numeric vector containing the conditional modes of the random effects.
- From the sim() output stored in s.out, you may extract quantities of
interest via getters:
- s.out$getqi(qi = "pv"): the simulated predicted values drawn from the distributions defined by the expected values.
- s.out$getqi(qi = "ev"): the simulated expected values for the specified values of x.
- s.out$getqi(qi = "fd"): the simulated first differences in the expected values for the values specified in x and x1.
- s.out$getqi(treatment): the simulated average predicted treatment effect for the treated from conditional prediction models.
See also¶
Mixed effects Poisson regression is part of lme4 package.
Bates D, Mächler M, Bolker B and Walker S (2015). “Fitting Linear Mixed-Effects Models Using lme4.” Journal of Statistical Software, 67 (1), pp. 1-48. doi: 10.18637/jss.v067.i01 (URL: http://doi.org/10.18637/jss.v067.i01).